Statistical models for dynamic networks
Presenter
March 1, 2012
Keywords:
- Systems biology; networks
MSC:
- 92C42
Abstract
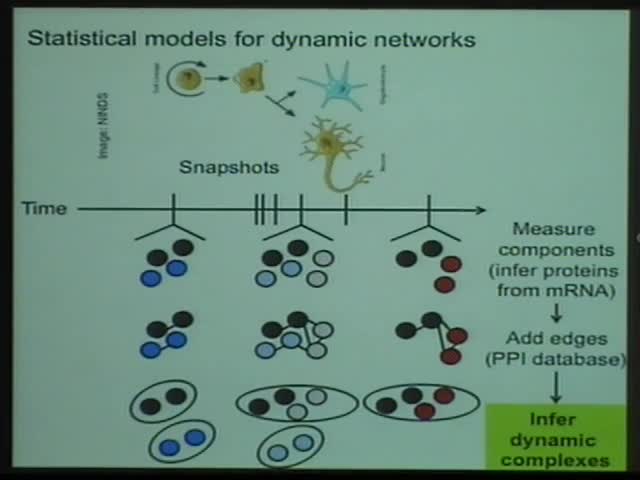
Biological networks are dynamic, driven by processes such as development and disease that change the expressed genes and proteins and modify interactions. Understanding how networks remodel could reveal the etiology of developmental disorders and suggest new drug targets for infectious disease. We present new methods that predict how networks change over time through joint analysis of time-domain and static data. For each single network snapshot, we infer a hierarchical stochastic block model that describes patterns of heterogeneous edges connecting inferred groups of vertices. Across snapshots we introduce a time-evolution operator similar to a path integral or hidden Markov model for group membership. The method is fully Bayesian, with no adjustable parameters. Our methods provide superior performance for predicting new interactions and protein complex co-membership. Applied to dynamic data, the method reveals new regulatory mechanisms for controlling the activity of multi-protein complexes. These methods may have broad applicability to social networks and other dynamic networks.
Previous related work:
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0008118 http://www.biomedcentral.com/1471-2105/12/S1/S44